Validation sub-mode
The validation sub-mode of MobyWat can be used for test and calibration of clustering algorithms implemented in Prediction mode and also in search for optimal molecular dynamics simulation parameters (Jeszenői 2016b). Validation sub-mode is mostly based on the algorithms of Prediction mode. It allows a comparison with a reference water positions for calculation of the rate of successful predictions at different clustering schemes, tolerances, and simulation parameters. Validation sub-mode can be recommended for testing new force fields and explicit water models (Jeszenői 2016b).
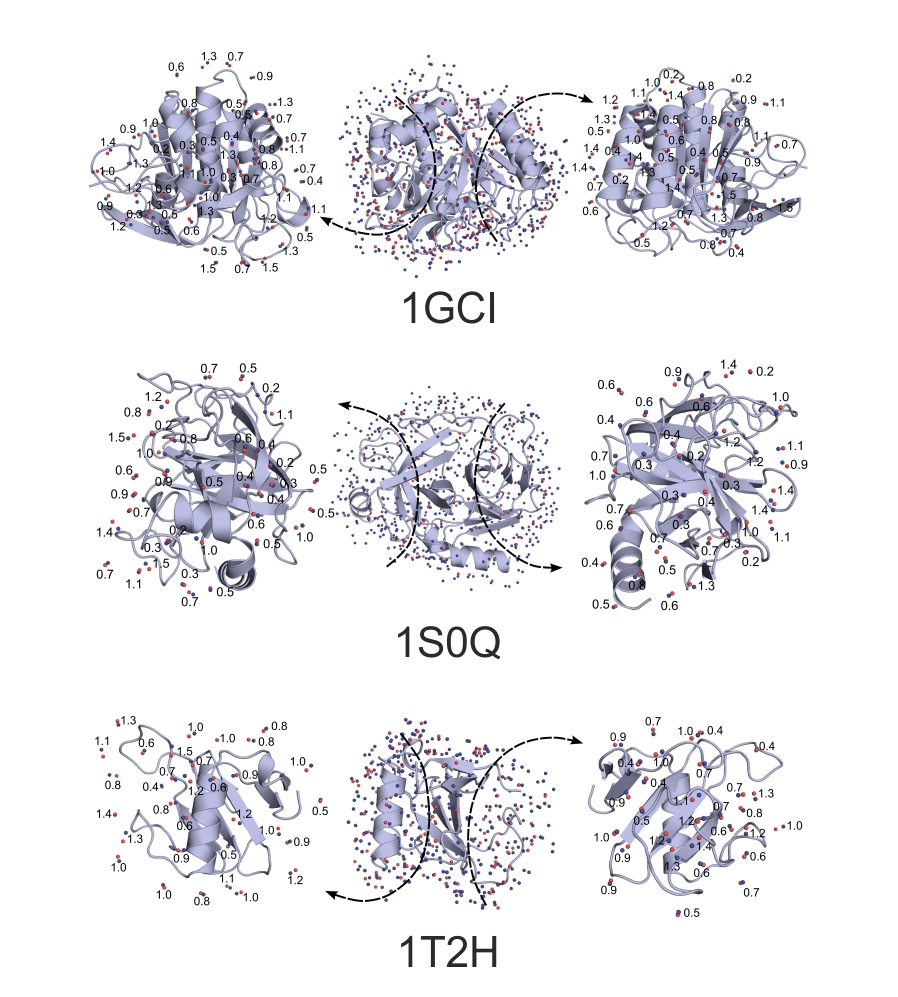
Predicted water positions (blue spheres) of three protein systems matching with crystallographic reference water positions (red spheres). 83.9, 83.7, and 79.0 % of the crystallographic reference positions were found, respectively. PDB codes are shown. Matching pairs and distances (Å) are shown for clarity. Results on additional protein systems were also published (Jeszenői et al. 2015).
A detailed User's Manual and test packages for Validation sub-mode can be found on the Download page