Program details
1 Organization of code
MobyWat was written in standard C, and therefore, the source code shares known benefits of the language such as portability and efficient dynamic memory allocation. Thus, introduction of theoretical limit on system size was not necessary, which is beneficial for atomic systems of large target molecules of various sizes and up to tens of thousands of water molecules surrounding them in the simulation box. A schematic organization chart of the program is given in Fig. PD1.1 as an overview of the main connections and I/O data flow in the code.

Fig. PD1: Main connections and I/O flow in MobyWat source code
2 Water pool data type
MobyWat uses a pool list with spatial coordinates, atom- and residue serial numbers of oxygen atoms for identification of a water molecule in the pool (Fig. PD2). The binary pool waters file stores pool lists of all selected waters and all frames specified at command line. Besides the benefits of shorter repeated runs, creation of plw files allows efficient use of disk space and memory.
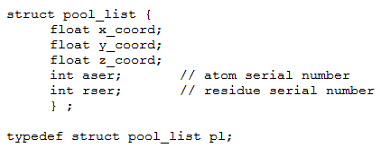
Fig. PD2: Water pool data type
3 Binary trajectory files
At present, MobyWat handles xtc files. GROMACS xtc files are based on extended xdr libraries written using an algorithm where precision can be set by scale factor, typically 1000 used for multiplication of the coordinates in nm and the values are rounded as integers. The modified xdr libraries were adopted by MobyWat for reading and writing xtc files, and the precision was increased to 10000 to handle PDB coordinates in Å from any source. The modified xdr libraries and their documentation can be found here.
