Glossary
Frame. Mobility of water molecules can be followed at an atomic scale by calculation methods such as molecular dynamics. As a result of a molecular dynamics run, spatial positions of all atoms are recorded at regular time steps. The set of coordinates of all atoms recorded after a time step is called a frame.
Hydration types. MobyWat predicts hydration of the surface (SF) of a single solute molecule (target) or the interface (IF) of a target-ligand complex. Tests of SF predictions were published (Jeszenői et al. 2015a). Results of experimenting on extra tricks for “filling” the IF region with water and the corresponding methodology will be submitted for publication soon (Jeszenői et al. 2015b). Fig. G1 shows molecular situations for both hydration types.

Fig. G1: Hydration types
Reference structure. Experimental structure of a target or a target-ligand complex with the surrounding water molecules. Most of reference structures are produced by crystallography where water molecules are represented by their oxygen atoms. See also water pools.
Solute. A target (usually a large molecule) and optionally a ligand (usually a small molecule) surrounded by water or other solvent.
Trajectory. A joint collection (log-book) of a time-series of frames. A trajectory includes all four dimensional mobility information on water molecules including three Cartesian coordinates along the time dimension. Trajectories can be stored as separate or NMR-type PDB files or, preferably, in portable binary files.
Water pools. Frames and reference structures include all water molecules at various distances from the solute. Importantly, during the generation of the frames molecular dynamics does not distinguish between bulk waters and others close to the solute molecule allowing continuous exchange of any water molecules with each-other in the system. For MobyWat evaluations it is reasonable to separate a pool of water molecules with possible structural role to distinguish them from bulk waters of no use. A maximal distance limit (dmax) is used for the distinction. In the case of surface hydration, a water molecule is selected for the reference or candidate pools if a distance (dT in Fig. G1) measured between its oxygen atom and the closest heavy atom of the target satisfies Eq. G1.
 Eq. G1
Eq. G1
MobyWat generates pools from experimental data (reference pool in analysis mode and validation sub-mode) or from frames of an MD trajectory (candidate pool in prediction mode and validation sub-mode) according to Fig. G2.
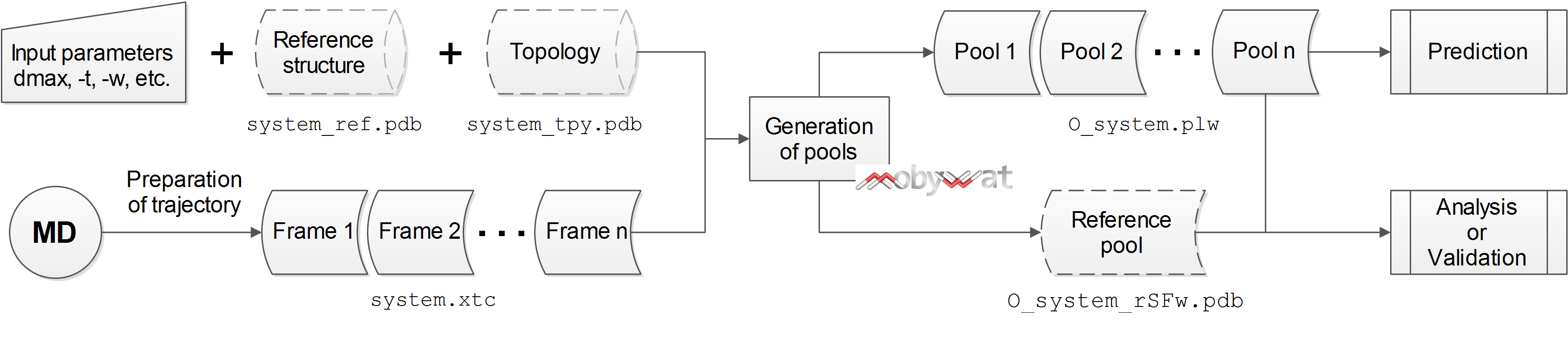
Fig. G2: Generation of water pools
